Wanderson de Sousa Mendes, Tomislav Hengl, Jonathan Sanderman, Katherine Todd-Brown, Leandro Leal Parente, Asa Gholizadeh
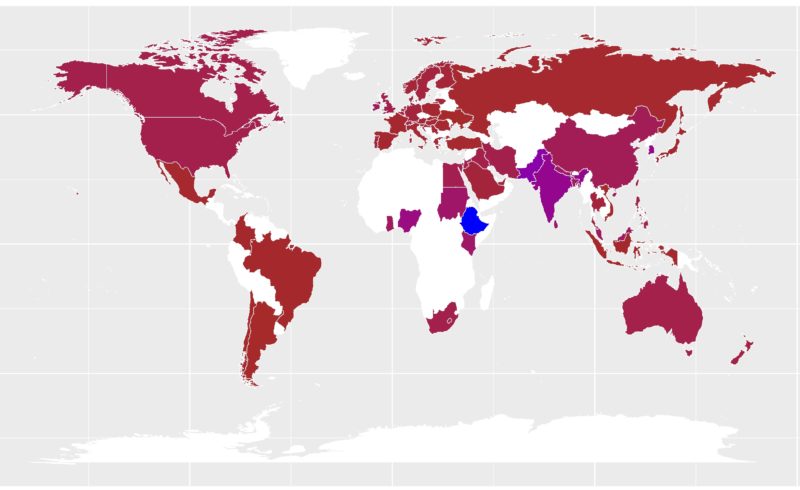
Essentially, the soil spectroscopy (SoilSpec) refers to the detection and analysis of the interaction between soil constituents and light (e.g. electromagnetic radiation) mainly across the (i) visible (Vis, 350 – 780 nm), (ii) near-infrared (NIR, 780 – 2500 nm), and (iii) mid-infrared (MIR, 2500 – 25000 nm or 4000 – 400 cm-1) spectral region. The methods of SoilSpec provide quick soil analysis at a low-cost and are environmentally-friendly way (no hazardous components are used in the analysis) compared to conventional laboratory analysis. The first study in this field of soil science can be traced back to the work of Stoner and Baumgardner (1981), who analysed the characteristic variation in reflectance of 485 soil samples from Brazil and the United States. Since then, research on SoilSpec has shown an upward trend (Nocita et al., 2015; Stenberg et al., 2010). A simple search into the Google Trends from 2004 – 2020 showed that SoilSpec has been widely spread (Fig. 1).
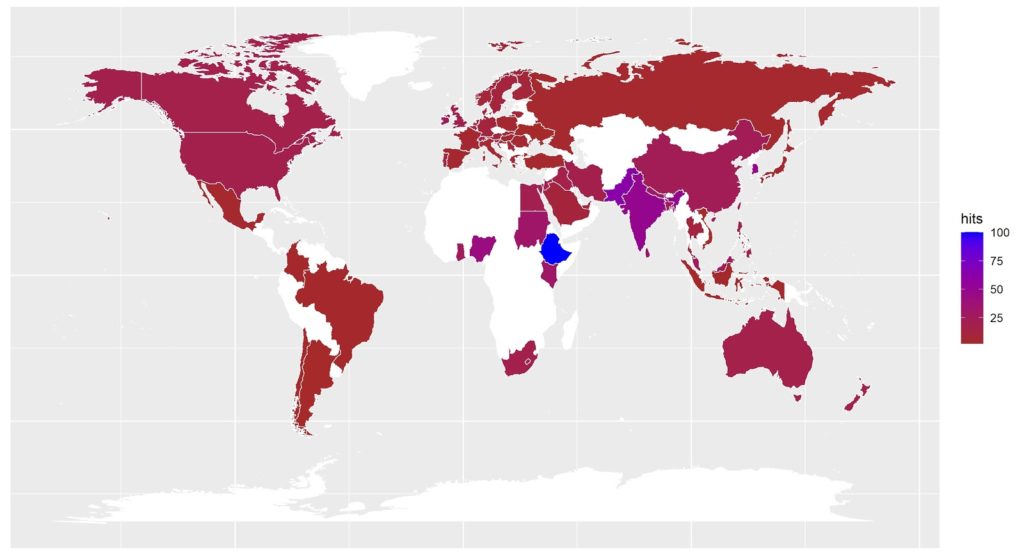
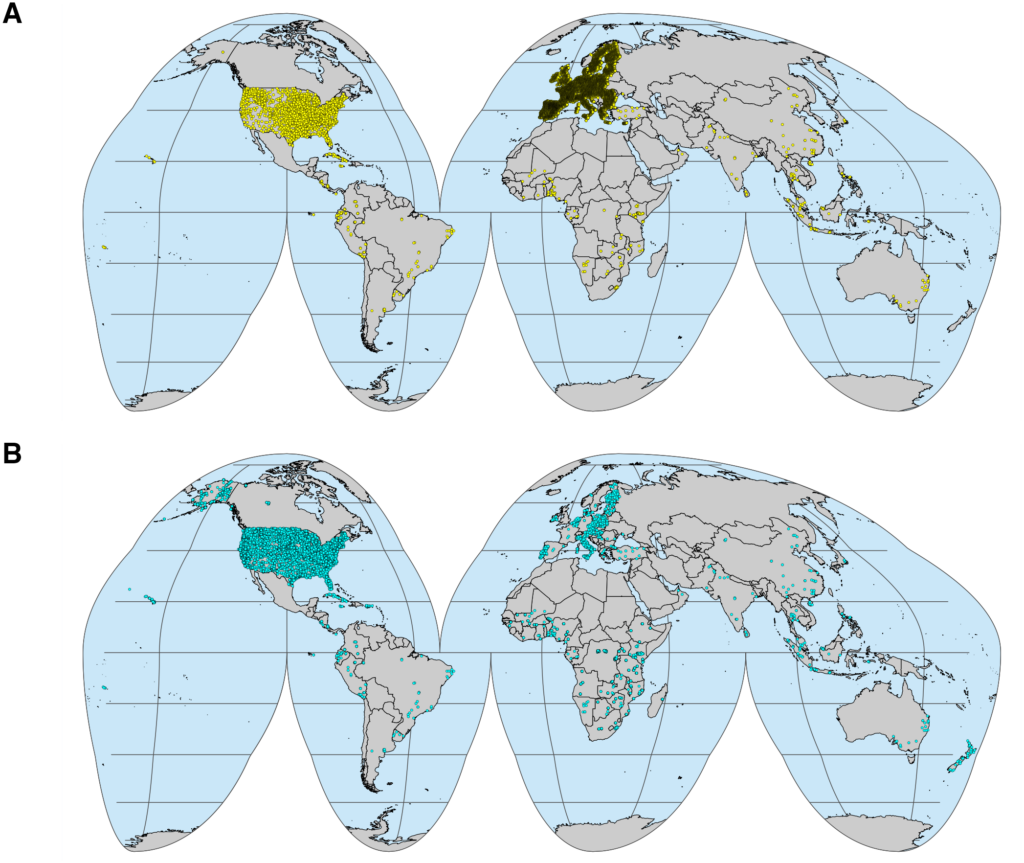
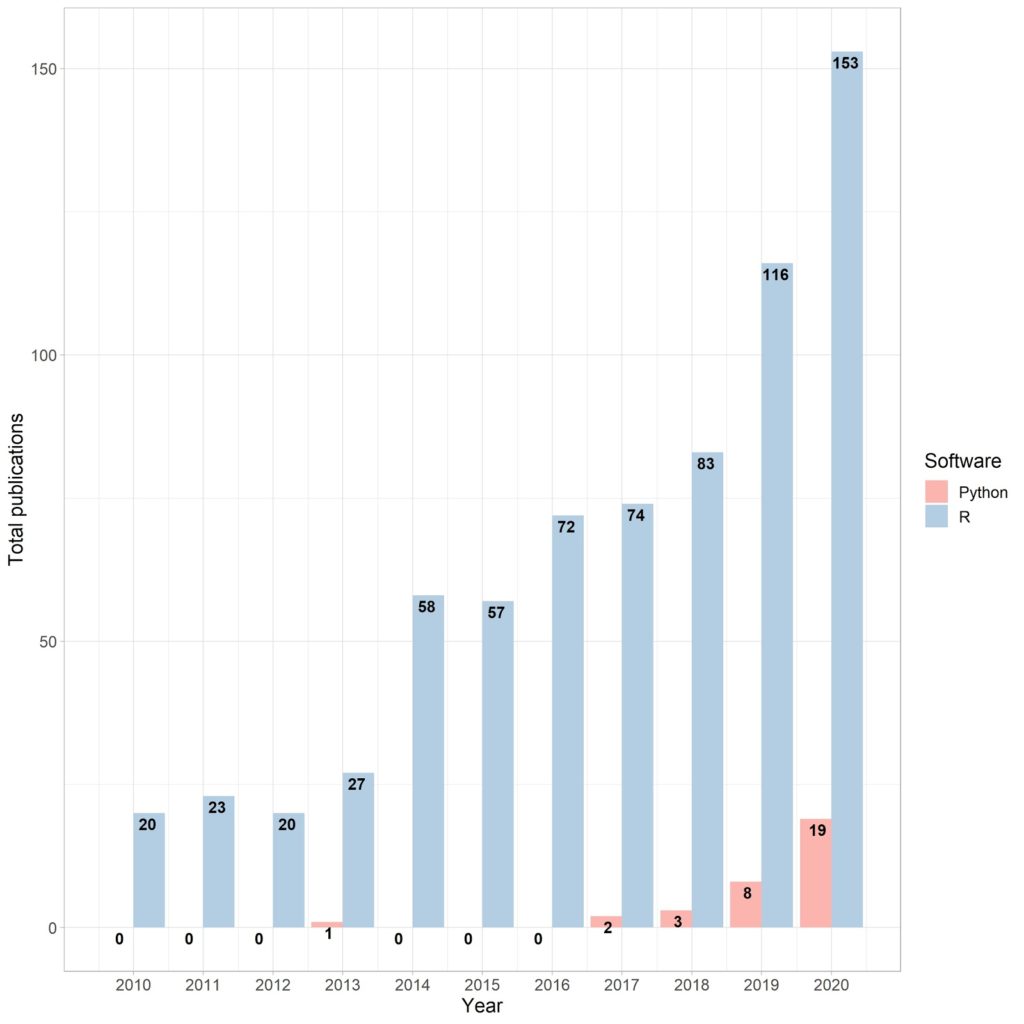
Implementation of SoilSpec methods in the open-source software programs has allowed soil scientists to solve economic, environmental, and health issues related to soil complexity using a massive volume of data, which is also considered as the main aim of the Soil Spectroscopy 4 Global Good (SoilSpec4GG) project. The number of research papers in SoilSpec using open-source programs has shown an upward trend (Fig. 2). The advantages of open-source software programs consist of free and easy access, personalisation, and transparency for any user. In this sense, the R (R Development Core Team, 2020), Python (Python Software Foundation, 2020), and Julia (Bezanson et al., 2017) programming languages are the key open-sources used for research in data and soil sciences. The first one is the most popular in soil spectroscopy because of the feasibility of packages for preprocessing, resampling, calibration sampling, and modelling (Table 1). Python and Julia have recent packages that work exactly the same as the “prospectr” R package. There are although, some specific differences among those programming languages. For instance, Julia is specifically designed to implement matrix expressions and linear algebra, which are the basis of data science. These features make Julia’s execution time faster than Python and R. However, R and Python have a huge set and number of libraries for SoilSpec compared with Julia. Moreover, the large community of R and Python serves an enormous benefit for developers. Most of the packages available for SoilSpec can be found in R programming, but new packages have been released using Python and Julia.
The open-source libraries deal with all peculiarities encountered in the soil spectral data, that are signal harmonisation, processing, calibration, and validation for modelling (Table 1 and Fig. 3). The preprocessing is the first step and consists in improving spectral response quality before modelling. If the soil spectral data were acquired from different sensors, it has to be resampled in order to harmonise the dataset. There are several preprocessing methods available, but Savitzky-Golay smoothing and derivative (Savitzky and Golay, 1964), continuum removal (Clark and Roush, 1984), and standard normal variate (Barnes et al., 1989) are the most suitable in dealing with Vis–NIR–MIR diffuse reflectance. These mathematical procedures improve data quality and reproducibility before predictive modelling. Then, the preprocessed soil spectral data have to be randomly split into calibration and validation sets. This procedure is crucial to avoid the effects of spectral autocorrelation and model bias. The calibration set is used to train the models whilst the independent validation serves to evaluate the fitness of the calibrated models. Afterwards, the modelling framework is the final step, wherein the algorithm is chosen and modelled using the calibration set. The soil spectral data modelling is performed manipulating machine learning algorithms such as cubist, random forest (RF), memory-based learning (MBL), partial least squares regression (PLSR), support vector machine (SVM), and convolutional neural network (CNN) (Dangal et al., 2019; Deiss et al., 2020; Ng et al., 2020, 2019; Stenberg et al., 2010).
| Software | Packages | Functionality | References |
|---|---|---|---|
| Preprocessing | |||
| R | prospectr | Signal processing Resampling | (Stevens and Ramirez-Lopez, 2020) |
| Python | nippy | Signal processing Resampling | (Torniainen et al., 2020) |
| Julia | Spectra | Signal processing Resampling | (Le Losq, 2016) |
| Sampling | |||
| R | caret | Calibration | (Kuhn, 2008) |
| prospectr | Calibration | (Stevens and Ramirez-Lopez, 2020) | |
| Clustering | |||
| R | ppclust | Spectral clustering | (Cebeci et al., 2020) |
| Spectrum | Spectral clustering | (John et al., 2019) | |
| Modelling | |||
| R | resemble | Memory-based learning | (Ramirez‐Lopez et al., 2020) |
| Cubist | Quinlan’s M5 model tree | (Kuhn and Quinlan, 2013) | |
| e1071 | Support Vector Machine | (Dimitriadou et al., 2008) | |
| randomForest | Random Forests | (Liaw and Wiener, 2002) | |
| pls | Partial least squares regression | (Mevik et al., 2011) | |
| kerasR | Convolutional Neural Network | (Arnold, 2017) | |
| Python | scikit-learn | Machine learning | (Pedregosa et al., 2011) |
| Julia | Spectra | linear and nonlinear programming | (Le Losq, 2016) |
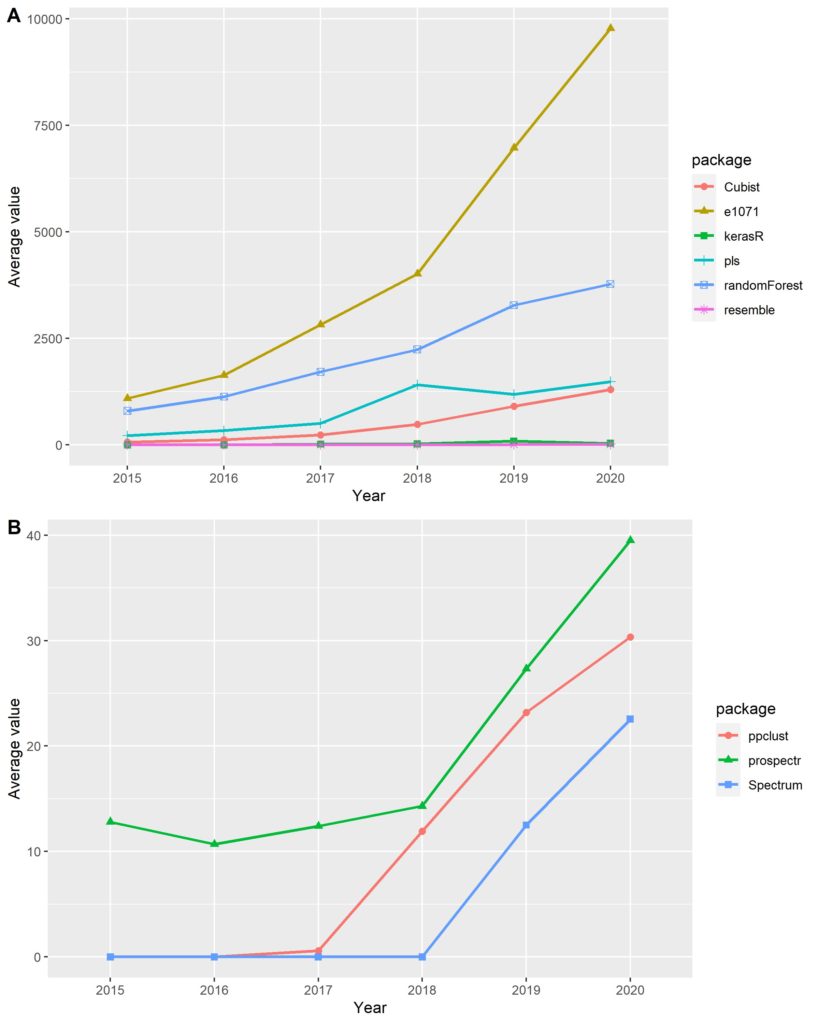
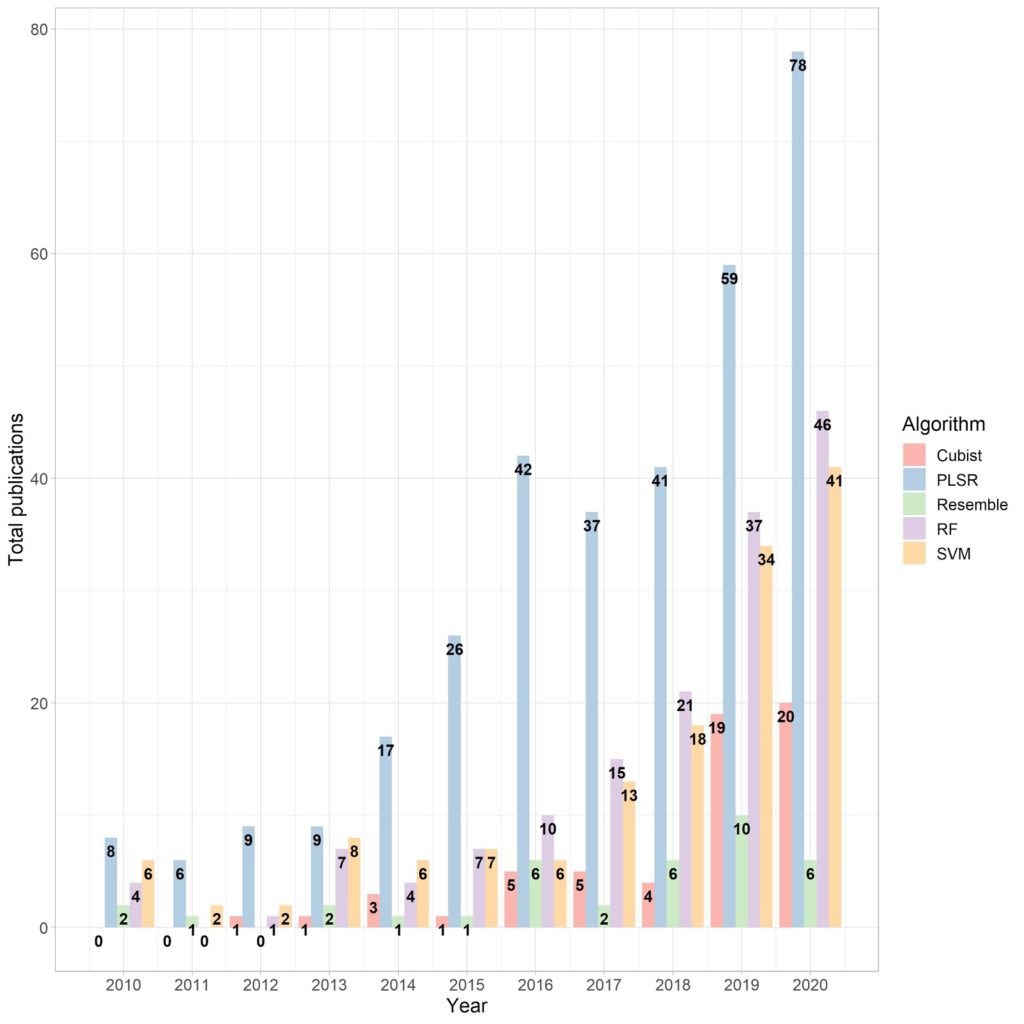
Fig. 4 illustrates the total number of academic articles for the five outstanding machine learning (ML) frameworks applied in SoilSpec between 2010 – 2020. Why are they preferable to other models? It is because these ML algorithms can deal with intricate nonlinear interactions between the predictor and response variables (Dangal et al., 2019). The first and most used method for predicting soil attributes using spectral predictors was the PLSR, which is a linear multivariate regression model including principal components and multiple linear regression. The preference for PLSR is due to its capacity of handling data with a large number of predictors with high collinearity. However, its application has shown a downward trend since the ML algorithms have proved to improve modelling performance for nearly all of the soil attributes. The cubist plays creating one or more rules for each partition with similar spectral characteristics and whether the rule is met, the linear regression of that partition is applied to create the prediction. The CNN allows the prospect of multi-task learning and the case of fusing inputs from unlike sources in diverse ways. This capacity can be helpful integrating for example vis-NIR and MIR spectra. The SVM is a nonparametric, supervised and statistical learning method that tries to keep the equilibrium between generalised trained models and predictive performance to unseen data (Gholizadeh et al., 2013). The main benefits of SVM are its ability to deal with noisy patterns, multimodal distributions of soil attributes and spectra. The RF is an ensemble approach that applies decision trees to unfold regression and classification issues based on rules in each tree to binary split data. Handling spectral data, the RF has proved to overfit some predictions. That is why it has hit the lowest preference in SoilSpec. Last but not least, MBL uses multiple learning algorithms improving predictive models for the same increase in computer processing of other methods. Basically, it fits a target function using a small subset in order to predict a large set using for instance the correlation within spectra or the principal component distances. Therefore, there is no perfect prescription of which ML method has to be always performed in SoilSpec, but those five should be always considered in dealing with SoilSpec.
Promising papers in the literature have proved the efficiency of open-source libraries for SoilSpec. Nawar and Mouazen (2019) predicted soil organic carbon using an online Vis-NIR (370 – 1979 nm) spectra coupled with RF. The steps carried out by those authors were Savitzky-Golay (e.g. preprocessing/ prospectr R package) and RF modelling (randomForest R package). Another study compared the performance of CNN, PLSR, and cubist models using single Vis–NIR, MIR and combined Vis–NIR–MIR to predict some soil attributes (Ng et al., 2019). The soil spectral data were preprocessed using Savitzky-Golay smoothing followed by standard normal variate (prospectr R package). The PLSR and cubist models were implemented using pls and Cubist R packages and the CNN was implemented in Python (Keras library). Those studies proved the higher relevance of open-source software programs in SoilSpec.
In general, the current developments in SoilSpec are directly related to the availability of open-source programs and have increased rapidly as new packages are released. The future prospects for the practical implementation of SoilSpec are promising as global initiatives including SoilSpec4GG have emerged by accelerating developments in SoilSpec via a global collaborative open-source platform.
References
Arnold, T., 2017. kerasR: R Interface to the Keras Deep Learning Library [WWW Document]. R Package. URL https://cran.r-project.org/web/packages/kerasR/index.html
Barnes, R.J., Dhanoa, M.S., Lister, S.J., 1989. Standard Normal Variate Transformation and De-Trending of Near-Infrared Diffuse Reflectance Spectra. Appl. Spectrosc. 43, 772–777. https://doi.org/10.1366/0003702894202201
Bezanson, J., Edelman, A., Karpinski, S., Shah, V.B., 2017. Julia: A Fresh Approach to Numerical Computing. SIAM Rev. 59, 65–98. https://doi.org/10.1137/141000671
Cebeci, Z., Yildiz, F., Kavlak, A.T., Cebeci, C., Onder, H., 2020. ppclust: Probabilistic and Possibilistic Cluster Analysis [WWW Document]. R Package. Vignette. URL https://cran.r-project.org/web/packages/ppclust/index.html
Clark, R.N., Roush, T.L., 1984. Reflectance spectroscopy: Quantitative analysis techniques for remote sensing applications. J. Geophys. Res. Solid Earth 89, 6329–6340. https://doi.org/10.1029/JB089iB07p06329
Dangal, S., Sanderman, J., Wills, S., Ramirez-Lopez, L., 2019. Accurate and Precise Prediction of Soil Properties from a Large Mid-Infrared Spectral Library. Soil Syst. 3, 11. https://doi.org/10.3390/soilsystems3010011
Deiss, L., Margenot, A.J., Culman, S.W., Demyan, M.S., 2020. Tuning support vector machines regression models improves prediction accuracy of soil properties in MIR spectroscopy. Geoderma 365, 114227. https://doi.org/10.1016/j.geoderma.2020.114227
Dimitriadou, E., Hornik, K., Leisch, F., Meyer, D., Maintainer, A.W., 2008. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien [WWW Document]. R Package. URL https://cran.r-project.org/package=e1071 (accessed 10.27.18).
Gholizadeh, A., Luboš, B., Saberioon, M., Vašát, R., 2013. Visible, near-infrared, and mid-infrared spectroscopy applications for soil assessment with emphasis on soil organic matter content and quality: State-of-the-art and key issues. Appl. Spectrosc. https://doi.org/10.1366/13-07288.
John, C.R., Watson, D., Barnes, M.R., Pitzalis, C., Lewis, M.J., 2019. Spectrum: fast density-aware spectral clustering for single and multi-omic data. Bioinformatics 36, 1159–1166. https://doi.org/10.1093/bioinformatics/btz704
Kuhn, M., 2008. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 28, 1–26. https://doi.org/10.18637/jss.v028.i05
Kuhn, M., Quinlan, R., 2013. Cubist: rule-and instance-based regression modeling [WWW Document]. R Package. URL https://cran.r-project.org/web/packages/Cubist/index.html (accessed 12.3.20).
Le Losq, C., 2016. Spectra. jl: A Julia package for processing spectroscopic data [WWW Document]. https://doi.org/10.5281/zenodo.53940
Liaw, A., Wiener, M., 2002. Classification and Regression by randomForest. R News 2, 18–23.
Massicotte, P., Eddelbuettel, D., 2020. gtrendsR: Perform and Display Google Trends Queries. [WWW Document]. R Package. URL https://cran.r-project.org/package=gtrendsR (accessed 12.7.20).
Mevik, B.-H., Wehrens, R., Liland, K.H., 2011. pls: Partial Least Squares and Principal Component Regression [WWW Document]. R Package. URL https://cran.r-project.org/web/packages/pls/index.html (accessed 12.3.2020).
Nawar, S., Mouazen, A.M., 2019. On-line vis-NIR spectroscopy prediction of soil organic carbon using machine learning. Soil Tillage Res. 190, 120–127. https://doi.org/10.1016/j.still.2019.03.006
Ng, W., Minasny, B., Mendes, W. de S., Demattê, J.A.M., 2020. The influence of training sample size on the accuracy of deep learning models for the prediction of soil properties with near-infrared spectroscopy data. SOIL 6, 565–578. https://doi.org/10.5194/soil-6-565-2020
Ng, W., Minasny, B., Montazerolghaem, M., Padarian, J., Ferguson, R., Bailey, S., McBratney, A.B., 2019. Convolutional neural network for simultaneous prediction of several soil properties using visible/near-infrared, mid-infrared, and their combined spectra. Geoderma 352, 251–267. https://doi.org/10.1016/j.geoderma.2019.06.016
Nocita, M., Stevens, A., van Wesemael, B., Aitkenhead, M., Bachmann, M., Barthès, B., Ben Dor, E., Brown, D.J., Clairotte, M., Csorba, A., Dardenne, P., Demattê, J.A.M., Genot, V., Guerrero, C., Knadel, M., Montanarella, L., Noon, C., Ramirez-Lopez, L., Robertson, J., Sakai, H., Soriano-Disla, J.M., Shepherd, K.D., Stenberg, B., Towett, E.K., Vargas, R., Wetterlind, J., 2015. Soil Spectroscopy: An Alternative to Wet Chemistry for Soil Monitoring, in: Advances in Agronomy. pp. 139–159. https://doi.org/10.1016/bs.agron.2015.02.002
Pedregosa, F., Varoquaux, G., Michel, V., Thirion, B., 2011. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830.
Python Software Foundation, 2020. Python Language Reference [WWW Document]. URL http://www.python.org (accessed 5.16.20).
R Development Core Team, R., 2020. R: A Language and Environment for Statistical Computing. [WWW Document]. URL https://www.r-project.org (accessed 5.16.2020).
Ramirez‐Lopez, L., Stevens, A., Rossel, R.V., Lobsey, C., Wadoux, A., Breure, T., 2020. resemble: Regression and similarity evaluation for memory-based learning in spectral chemometrics. [WWW Document]. R Package. URL https://cran.r-project.org/web/packages/resemble/index.html (accessed 12.2.20).
Savitzky, A., Golay, M.J.E., 1964. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 36, 1627–1639. https://doi.org/10.1021/ac60214a047
Stenberg, B., Viscarra Rossel, R.A., Mouazen, A.M., Wetterlind, J., 2010. Visible and Near Infrared Spectroscopy in Soil Science, in: Sparks, D.L. (Ed.), Advances in Agronomy. Academic Press, pp. 163–215. https://doi.org/10.1016/S0065-2113(10)07005-7
Stevens, A., Ramirez-Lopez, L., 2020. An introduction to the prospectr package. [WWW Document]. R Package. URL https://cran.r-project.org/web/packages/prospectr/index.html (accessed 11.30.20).
Stoner, E.R., Baumgardner, M.F., 1981. Characteristic Variations in Reflectance of Surface Soils1. Soil Sci. Soc. Am. J. 45, 1161. https://doi.org/10.2136/sssaj1981.03615995004500060031x
Torniainen, J., Afara, I.O., Prakash, M., Sarin, J.K., Stenroth, L., Töyräs, J., 2020. Open-source python module for automated preprocessing of near infrared spectroscopic data. Anal. Chim. Acta 1108, 1–9. https://doi.org/10.1016/j.aca.2020.02.030